当前你的浏览器版本过低,网站已在兼容模式下运行,兼容模式仅提供最小功能支持,网站样式可能显示不正常。
请尽快升级浏览器以体验网站在线编辑、在线运行等功能。
1344:Tree Size Problem
题目描述
Trees are used to represent the evolutionary relationship of species. An evolutionary tree is a edge-weighted tree with each leaf representing one species. The distance between two leaves on the tree represents the dissimilarity of these two species. An important issue in computational biology is to construct the evolutionary tree from the observed dissimilarities.
Let N = {1..n}. An n*n matrix M is a metric over N if it is symmetric, nonnegative, and M[i, j] + M[j, k] >= M[i, k] for any 1<= i, j, k <=n (i.e., triangle inequality). A metric is a tree metric if it can be realized by a tree, i.e., there exists an edge-weighted tree T such that
1. the leaf set is N;
2. the weight of each edge is nonnegative;
3. and d(T, i, j) = M[i, j] for any i, j <= N, where d(T, i, j) is the shortest path length between i and j on the tree T.
For example, the following matrix is a tree metric. The corresponding tree is given in Figure 8.
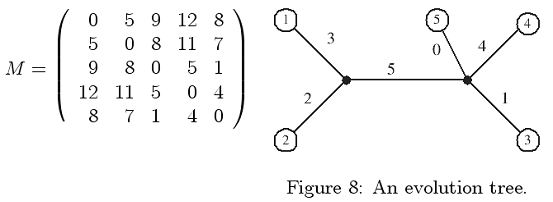
The size of a tree is defined to be the total weight of the tree edges. For a tree metric, it has been shown that the tree size is unique, i.e., it is impossible to find two trees of different sizes realizing the same tree metric. Your task is to design a program to compute the tree sizes of the given tree metrics. The following simple example may be helpful. For the case of only two species, the tree has only one edge and the tree size is just the distance between the two species. Let us consider the case of three species a, b, and c. Let T be the corresponding tree. Since T has three leaves, there is an internal node x. By definition, the path length d(T, a, b) = M[a, b]. Since x is a vertex on the path between a and b, all we need to do is to determine the weight (length) of edge (x, c). Let w(x, c) denote the weight of edge (x, c). We have
w(x, c) + w(x, a) = M[a, c],
w(x, c) + w(x, b) = M[b, c],
and w(x, a) + w(x, b) = M[a, b].
Therefore, w(x, c) = (M[a, c] +M[b, c] -M[a, b])/2.
Let N = {1..n}. An n*n matrix M is a metric over N if it is symmetric, nonnegative, and M[i, j] + M[j, k] >= M[i, k] for any 1<= i, j, k <=n (i.e., triangle inequality). A metric is a tree metric if it can be realized by a tree, i.e., there exists an edge-weighted tree T such that
1. the leaf set is N;
2. the weight of each edge is nonnegative;
3. and d(T, i, j) = M[i, j] for any i, j <= N, where d(T, i, j) is the shortest path length between i and j on the tree T.
For example, the following matrix is a tree metric. The corresponding tree is given in Figure 8.

The size of a tree is defined to be the total weight of the tree edges. For a tree metric, it has been shown that the tree size is unique, i.e., it is impossible to find two trees of different sizes realizing the same tree metric. Your task is to design a program to compute the tree sizes of the given tree metrics. The following simple example may be helpful. For the case of only two species, the tree has only one edge and the tree size is just the distance between the two species. Let us consider the case of three species a, b, and c. Let T be the corresponding tree. Since T has three leaves, there is an internal node x. By definition, the path length d(T, a, b) = M[a, b]. Since x is a vertex on the path between a and b, all we need to do is to determine the weight (length) of edge (x, c). Let w(x, c) denote the weight of edge (x, c). We have
w(x, c) + w(x, a) = M[a, c],
w(x, c) + w(x, b) = M[b, c],
and w(x, a) + w(x, b) = M[a, b].
Therefore, w(x, c) = (M[a, c] +M[b, c] -M[a, b])/2.
输入解释
The input file consists of several test cases. The first line of each test case is a positive integer n, 2 < n < 30. The following n - 1 lines represent the upper triangle of the tree metric, but the diagonal is not included. Each line is for one row, and elements are separated by spaces. All the elements are nonnegative integers less than 100. The last case is followed by a "0" to indicate the end of input. You may assume that the test data are all tree metrics, and it is not necessary to check them.
Furthermore, the size of a tree is the sum of all integers in the test case except the integers in the first line of the test case.
Furthermore, the size of a tree is the sum of all integers in the test case except the integers in the first line of the test case.
输出解释
For each test case, output the tree size in one line.
输入样例
5 5 9 12 8 8 11 7 5 1 4 4 15 36 60 31 55 36 0
输出样例
15 71
最后修改于 2020-10-29T06:01:30+00:00 由爬虫自动更新
共提交 0 次
通过率 --%
| 时间上限 | 内存上限 |
| 1000 | 10000 |
登陆或注册以提交代码