当前你的浏览器版本过低,网站已在兼容模式下运行,兼容模式仅提供最小功能支持,网站样式可能显示不正常。
请尽快升级浏览器以体验网站在线编辑、在线运行等功能。
3877:Genetics
题目描述
Alien DNA molecules have the structure of a circular sequence. Each sequence is composed of nucleotides. There are 26 different types of nucleotides, and each of them can occur in two faces. It is very important to remark that in any given alien DNA molecule, every nucleotide either does not appear at all or appears exactly twice (hence, the length of a DNA molecule is an even integer between 2 and 52). In case a nucleotide occurs twice, each occurrence can be of either type independently. Alien bacteria have two types of extremities, which in the technical biological jargon are referred to as arms and legs. A major discovery of Dr. Poucher's team is a method to determine the exact number of arms and legs of a bacterium by examining its DNA structure.
Here we represent each nucleotide as a letter of the alphabet. We refer to the different nucleotides as a, A, . . . z, Z, where the lowercase and uppercase forms of a letter represent the two possible faces a nucleotide may appear with; we shall also use a/A, b/B, . . . z/Z to refer to a nucleotide in either face. To determine the number of extremities, Dr. Poucher starts by initializing two counters of arms and legs to zero, and then proceeds to perform a number of surgeries, transforming a DNA sequence into another one. After each transformation, you may need to increase some of the counters, depending on the type of surgery applied. When the empty sequence of nucleotides (which will be denoted by __poj_jax_start__\emptyset__poj_jax_end__) has been reached, the number of extremities of the original molecule has been found. The possible surgeries are:
1. Eliminate consecutive instances of a given nucleotide appearing with opposite faces. The number of arms and legs is preserved. For example: aBbCaC -> aCaC by eliminating Bb. Another example: DeHhEd -> eHhE by eliminating dD. Remember that DNA structure is circular, so in our representation as a string the last and first letters are connected.
2. Eliminate consecutive nucleotides appearing with the same face. Add one to the number of arms. For example: BBcgCg -> cgCg by eliminating BB. Another example: xabyyaBX -> xabaBX by eliminating yy.
3. Eliminate a sequence of four nucleotides formed by two different nucleotides that appear alternately where different occurrences of the same nucleotide have opposite faces. Add one to the number of legs. For example: dcDCefFe -> efFe, by eliminating dcDC. Another example: cmNMnC -> cC by eliminating mNMn.
4. Cut and paste, the most sophisticated procedure. First, a nucleotide is selected, for instance a/A, and the DNA sequence is chopped into two linear chains such that the nucleotide appears once in each of them.
Second, if both occurrences of a/A are of the same face, one of the chains is "inverted" by reversing the sequence and changing the face of every nucleotide in the chain.
Then, the chains are combined by concatenating the subsequence occurring before a with the subsequence occurring after A, and the subsequence occurring after a with the sub-sequence occurring before A.
Finally, two new a/A nucleotides are added to close the chain into a circular shape. The face of the new nucleotides are the same if the original pair of nucleotides selected had the same face, and is different otherwise.
Formally, suppose you select the nucleotide a=A, and further assume for the moment that it appears both times with the face a (A). The cut and paste surgery turns sequences of the form __poj_jax_start__S_1aS_2S_3aS_4__poj_jax_end__ (respectively __poj_jax_start__S_1AS_2S_3AS_4__poj_jax_end__) into __poj_jax_start__S_2aS_1\overline{S}_3a\overline{S}_4__poj_jax_end__ (respectively __poj_jax_start__S_2AS_1\overline{S}_3A\overline{S}_4__poj_jax_end__). On the other hand, if nucleotide a\A appears with its two different faces, the surgery turns sequences of the form __poj_jax_start__S_1aS_2S_3AS_4__poj_jax_end__ into __poj_jax_start__S_2aS_1S_4AS_3__poj_jax_end__. S1, S2, S3 and S4 are arbitrary sub-chains (possibly empty). In both cases the original circular chain was chopped into __poj_jax_start__S_1(a/A)S_2__poj_jax_end__ and __poj_jax_start__S_3(a/A)S_4__poj_jax_end__.
For example (see the figure below): starting with the sequence BacDcAbD, we can get chains BacDc and AbD. Then, merging at nucleotide a=A we get the sequence cDca'BbDA' where a' and A' represent the new a/A nucleotides. Here, S1 = B, S2 = cDc, S3 = __poj_jax_start__\emptyset__poj_jax_end__; and S4 = bD.
Another example: take the same DNA sequence BacDcAbD, and cut to get the chains DBac and DcAb; paste nucleotide c/C (in this case you need to reverse one chain, for example BaCd) to get the sequence cDBadcBa. Here, S1 = DBa, S2 = __poj_jax_start__\emptyset__poj_jax_end__, S3 = D and S4 = Ab.
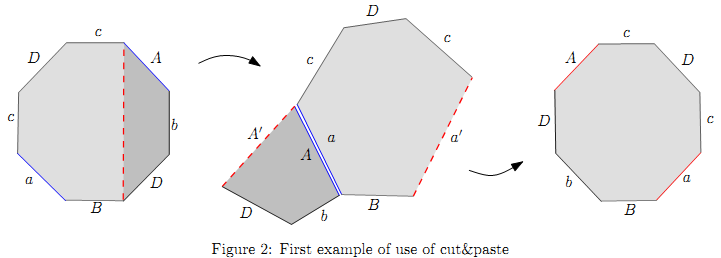
This surgery does not modify the number of arms or legs, but can be used cleverly in combination with the previous surgeries to reduce the size of the DNA molecule and finish the calculation.
However, alien bacteria do not present both arms and legs at the same time. This is due to the fact that, in their early development, a leg, in the presence of one or more arms, becomes two arms. Because of the above, the end result is either a number of arms or a number of legs, but not both at the same time. In order to avoid expensive surgical procedures, Dr. Poucher has hired you to write a program that computes the number of arms and legs a bacterium will develop, given its DNA sequence. It is guaranteed that the result is determined uniquely by the original string, regardless of the particular sequence of surgeries applied.
输入解释
输出解释
输入样例
rkrk abcdeABCDE shcoOCfFHS END
输出样例
1 arm 2 legs none
最后修改于 2020-10-29T07:14:16+00:00 由爬虫自动更新
共提交 0 次
通过率 --%
| 时间上限 | 内存上限 |
| 1000 | 65536 |