当前你的浏览器版本过低,网站已在兼容模式下运行,兼容模式仅提供最小功能支持,网站样式可能显示不正常。
请尽快升级浏览器以体验网站在线编辑、在线运行等功能。
2520:DNA Sequence Alignment
题目描述
Gnaileux Iew is attracted in Bioinformatics recently. He reads papers day and night and devotes all his mind in studying. Today he is going to review the basic problem in Bioinformatics: DNA sequence alignment. His purpose is to find a simple and effective algorithm that performs global alignment with two highly similar DNA sequences.
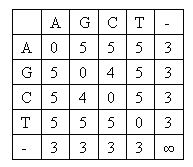
A DNA sequence is presented as a sequence of characters, which may be 'A', 'G', 'C' or 'T'. To align two DNA sequences, some gaps may be inserted to sequences so that two sequences have the same length. And then it is counted up for every pair of matched characters by a score matrix. Gnaileux Iew uses a minimal-score matrix hence the total score of alignment should be minimized. Following is the score matrix Gnaileux Iew uses:
For example, an alignment for DNA sequences "AAGACG" and "CAGAGCTC" may be:
-AAGA-C-G
CA-GAGCTC
The total score is 3+0+3+0+0+3+0+3+4=16.
Gnaileux Iew is only interested in aligning highly similar sequences. Strictly speaking, |LCS(A,B)| * 2 / (|A |+ |B|) >= 90%, where A and B are the sequences to align, and LCS(A,B) is the longest common subsequence of A and B.
A DNA sequence is presented as a sequence of characters, which may be 'A', 'G', 'C' or 'T'. To align two DNA sequences, some gaps may be inserted to sequences so that two sequences have the same length. And then it is counted up for every pair of matched characters by a score matrix. Gnaileux Iew uses a minimal-score matrix hence the total score of alignment should be minimized. Following is the score matrix Gnaileux Iew uses:

For example, an alignment for DNA sequences "AAGACG" and "CAGAGCTC" may be:
-AAGA-C-G
CA-GAGCTC
The total score is 3+0+3+0+0+3+0+3+4=16.
Gnaileux Iew is only interested in aligning highly similar sequences. Strictly speaking, |LCS(A,B)| * 2 / (|A |+ |B|) >= 90%, where A and B are the sequences to align, and LCS(A,B) is the longest common subsequence of A and B.
输入解释
Input contains multiple test cases. Each test case contains two lines, which are the two DNA sequences to align. DNA sequences contain only characters 'A', 'G', 'C' and 'T'. The length of each sequence is not greater than 50000.
You can assume that all the input cases are highly similar sequences.
You can assume that all the input cases are highly similar sequences.
输出解释
For each test case print the minimal total score of alignment in one line.
输入样例
AGTGCTGAAAGTTGCGCCAGTGAC AGTGCTGAAGTTCGCCAGTTGACG CACAATTTTTCCCAGAGAGA CGAATTTTTCCCAGAGAGA
输出样例
12 7
来自北京大学POJ的附加信息
| Case time limit(单组数据时间限制) | 5000MS |
最后修改于 2020-10-29T06:34:44+00:00 由爬虫自动更新
共提交 0 次
通过率 --%
| 时间上限 | 内存上限 |
| 25000 | 65536 |
登陆或注册以提交代码