当前你的浏览器版本过低,网站已在兼容模式下运行,兼容模式仅提供最小功能支持,网站样式可能显示不正常。
请尽快升级浏览器以体验网站在线编辑、在线运行等功能。
2941:Motif Finding
题目描述
Cell differentiation and development are fundamentally controlled by gene regulation. Only a subset of genes in the genome is expressed in a cell at a given time under given conditions. Regulatory sites on DNA sequence normally correspond to shared conservative sequence patterns among the regulatory regions of correlated genes. We call these conserved sequence motifs. The actual regulatory DNA sites corresponding to a motif are called the instances of that motif.
Identifying motifs and corresponding instances are very important, so biologists can investigate the interactions between DNA and proteins, gene regulation, cell development and cell reaction.
Given two equal-length strings, the Hamming Distance is the number of positions in which the corresponding characters are different. For example, the Hamming Distance between “ACTG” and “ATCG” is 2 because they differ at the 2nd and 3rd positions.
Your task is to find a motif for a few DNA sequences. Every DNA sequence consists of only A, C, G, T. If a substring of DNA sequence S has the same length with motif P and their Hamming Distance is not more than d, we say that S includes an instance of P. Given n DNA sequences with length of l. Among them, m sequences are “key sequences”. You need to find out the motif P whose length is w, so that every key sequence includes an instance of motif P. What’s more, the number of DNA sequences which include an instance of P should be as large as possible.
For example, n=7, m =7, l=40, w =8, d =2. The sequences are as follows (they are all key sequences):
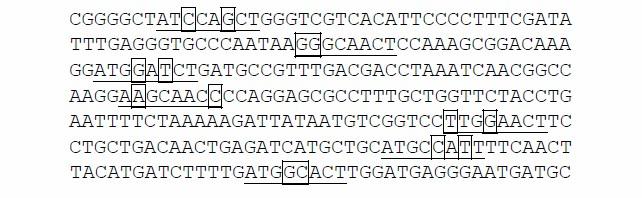
The motif P is ATGCAACT.
Identifying motifs and corresponding instances are very important, so biologists can investigate the interactions between DNA and proteins, gene regulation, cell development and cell reaction.
Given two equal-length strings, the Hamming Distance is the number of positions in which the corresponding characters are different. For example, the Hamming Distance between “ACTG” and “ATCG” is 2 because they differ at the 2nd and 3rd positions.
Your task is to find a motif for a few DNA sequences. Every DNA sequence consists of only A, C, G, T. If a substring of DNA sequence S has the same length with motif P and their Hamming Distance is not more than d, we say that S includes an instance of P. Given n DNA sequences with length of l. Among them, m sequences are “key sequences”. You need to find out the motif P whose length is w, so that every key sequence includes an instance of motif P. What’s more, the number of DNA sequences which include an instance of P should be as large as possible.
For example, n=7, m =7, l=40, w =8, d =2. The sequences are as follows (they are all key sequences):
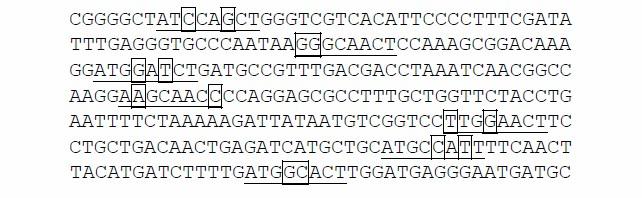
The motif P is ATGCAACT.
输入解释
The input contains several cases. The first line of each case contains three integers n (1 ≤ n ≤ 15), m (0 ≤ m ≤ n) and l (1 ≤ l ≤ 10,000). The second line contains two integers w (1 ≤ w ≤ 8) and d (0 ≤ d ≤ w). The third line contains m unique integers ranging from 0 to n-1, indicating the key sequences.
Then followed by n lines, in which each line contains one sequence. The input is terminated by three zeros.
Then followed by n lines, in which each line contains one sequence. The input is terminated by three zeros.
输出解释
For each case, output the motif P in one line. If the solution is not unique, then output the lexicographically smallest one. If there is no answer, you should output “No solution”.
输入样例
3 2 4 3 1 0 2 ACTT CGTG CCCC 3 2 4 3 0 0 2 ACTT CGTG CCCC 0 0 0
输出样例
CCT No solution
来自杭电HDUOJ的附加信息
| Recommend | lcy |
最后修改于 2020-10-25T22:58:26+00:00 由爬虫自动更新
共提交 0 次
通过率 --%
| 时间上限 | 内存上限 |
| 5000/2000MS(Java/Others) | 32768/32768K(Java/Others) |
登陆或注册以提交代码